EpiDirect® MLH1 Methylation qPCR Assay
The EpiDirect® MLH1 Methylation qPCR Assay represents a new and innovative approach allowing for direct quantification of MLH1 promoter methylation by qPCR without prior DNA conversion.
The EpiDirect® technology is based on our unique INA® chemistry and includes an EpiPrimer™ oligonucleotide that specifically binds and primes amplification of methylated target DNA.
The assay is able to quantify MLH1 promoter methylation using standard real-time PCR instruments* in just two hours with as little as 0.5 ng of unconverted input DNA.
- Standard qPCR instrument procedure
- No need for prior conversion of DNA
- Results in less than 2 hours
- Minimal hands-on time
- 0.5-50 ng input DNA
- For Research Use Only (RUO)
- CE IVDR expected in Q4 2025
*Validated on CFX96/CFX Opus 96 (Bio-Rad), QuantStudio™5 (ABI) and BaseTyper™ 48.4 (PentaBase) Real-Time PCR Systems
*Will be available from August 15th
Product Details
Reference Number 8400
Product Name EpiDirect® MLH1 Methylation qPCR Assay
Product Description PCR master mix and oligonucleotide mix for quantification of MLH1 promoter methylation, positive and negative control
Format Ready-to-Use
Reactions 48
Intended Use
EpiDirect® MLH1 Methylation qPCR Assay is a semi-automated real-time Polymerase Chain Reaction (PCR) assay intended for the direct, qualitative analysis of the methylation status of the human mismatch repair gene promotor MutL Homolog1 (MLH1).
The assay is intended for purified genomic DNA (gDNA) samples that, without further pre-treatment, can be used with real-time PCR systems to detect and quantify the methylation status of MLH1 promotor. Samples comprise Formalin-Fixed Paraffin-Embedded (FFPE) and fresh frozen biopsies and can be prepared using automated platforms or in manual workflows.
The assay is intended for use by health professionals or qualified laboratory personnel specifically instructed and trained in the techniques of Real-Time PCR as well as proficient in handling biological samples.
Product Specifications
Procedure Real-time PCR-based
Validated Real-Time PCR instruments BaseTyper™ 48.4 Quiet HRM (PentaBase Ref. No. 754), CFX96 and CFX Opus 96 (Bio-Rad) and QuantStudio™ 5 Real-Time PCR Systems
PentaBase technologies EpiPrimer™, HydrolEasy® Probe, SuPrimer™
Genomic target CpG sites 1-4 in MLH1 promoter Region C (-248 to -178)
Sample types Solid colorectal and endometrial cancer biopsies including formalin-fixed paraffin-embedded (FFPE) tissue samples
Sample tumor cell percentage At least 20%
Sample DNA extraction methods Relevant commercially avaliable solid biospy/FFPE DNA extraction kits
Sample input 0.1-10 ng/µL of purified DNA directly from the sample that has not been otherwise chemically manipulated
PCR run time Less than 2 hours
Instructions For Use
The latest version of the Instructions For Use: 5ba.se/ED002_RUO
Publications
A qPCR technology for direct quantification of methylation in untreated DNA
Bendixen et al. – Nature Communications – 2023
Related files
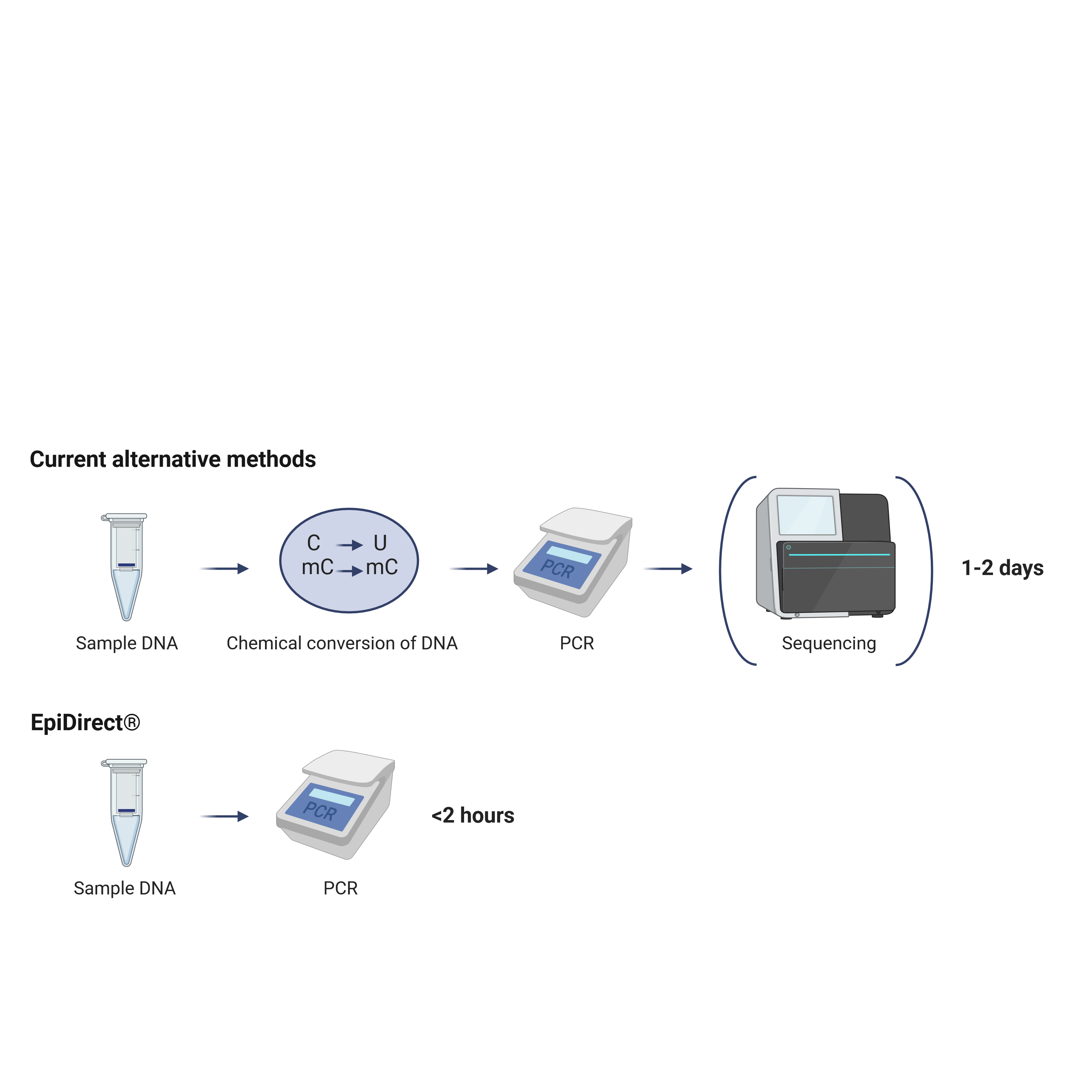
A Revolutionary Approach to DNA Methylation Testing
Current methods for evaluating DNA methylation are relying on sodium bisulfite or other chemical or enzymatical conversion of cytosines. However, variability in conversion rates as well as DNA degradation resulting from chemical treatment may lead to false or in-conclusive results. Especially bisulfite treatments have been shown to lead to a biased degradation of unmethylated DNA and consequently an overestimation of the methylation frequency.
In contrast, the EpiDirect® technology allows for direct quantification of DNA methylation by PCR without any prior chemical or enzymatic treatment. This revolutionizing technology not only eliminates artefacts related to incomplete conversion of cytosines and DNA degradation but also vastly reduces hands-on-time and time to result.
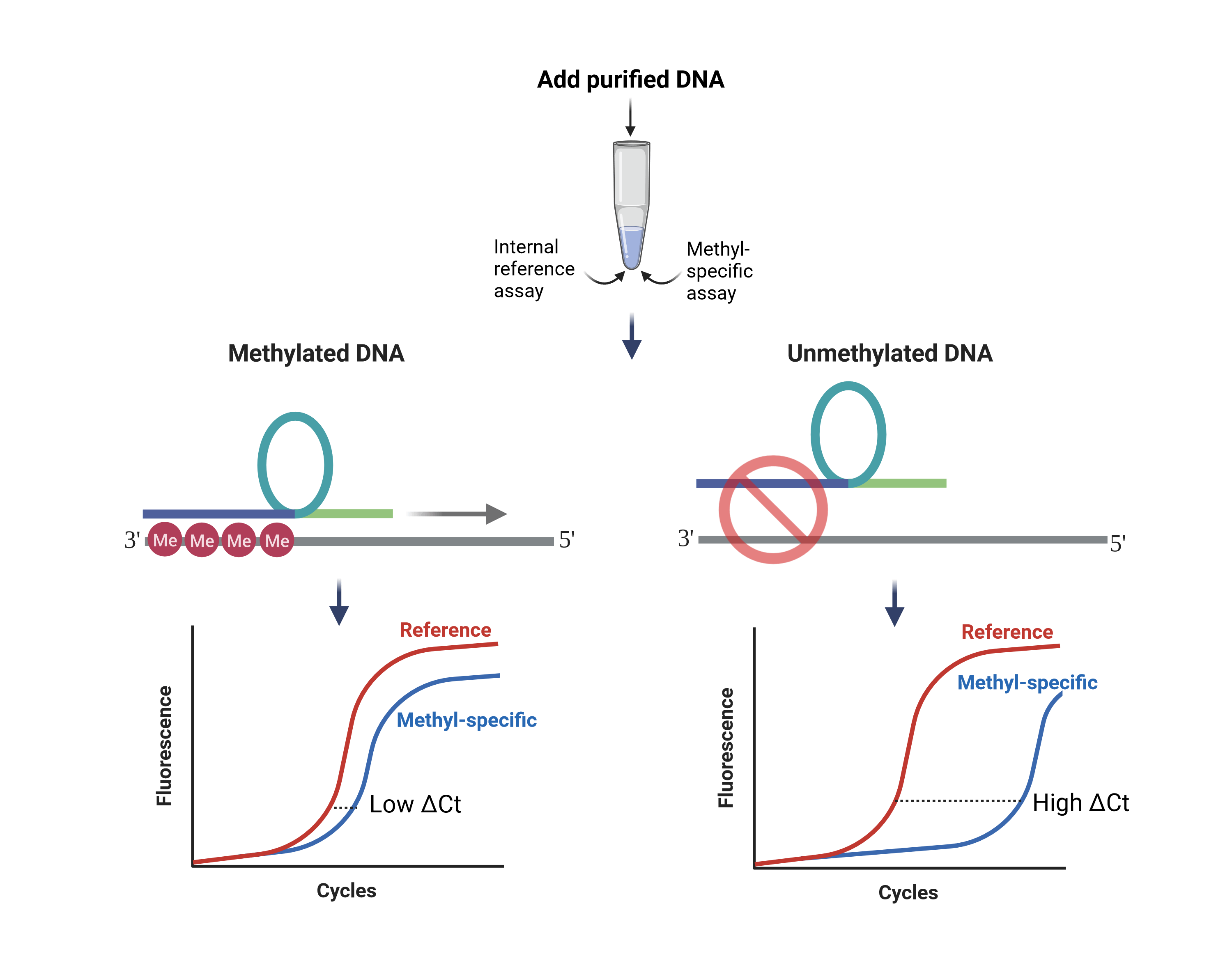
The EpiDirect® Technology
The EpiDirect® technology is based on a unique DNA analogue chemistry known as intercalating pseudo-nucleotide (IPN). IPN-containing oligonucleotides, also known as intercalating nucleic acids (INAs®), show highly improved target affinity and specificity compared to standard chemistry oligonucleotides.
The EpiDirect® technology utilizes a type of INA® known as an EpiPrimer™ that selectively binds to and primes amplification of methylated DNA. The EpiPrimer™ consists of 3 parts: a) an IPN-containing anchor sequence that binds to the methylated target region, b) a starter sequence that primes DNA replication, and c) a loop sequence that is part of the subsequent methyl-unspecific priming. Thus, the anchor and starter sequence prime the initial replication of the methylated DNA while the loop and starter sequence combined work as a primer in the remaining PCR cycles.