EpiPrimers™
The INA®-based EpiPrimer™ enables direct evaluation of DNA methylation by qPCR without prior conversion of the sample such as chemical bisulfite treatment. Assays based on the EpiPrimer™ technology also require much lower amounts of sample DNA compared to methods using converted DNA.
When combined with PentaBase’s HydrolEasy® probe and SuPrimer™, the EpiPrimer™ technology can be used to design powerful customised EpiDirect® qPCR assays that directly measure the methylation frequency of relevant targets.
- Low sample DNA input requirements
- Can be customised to specific targets
- No need for chemical conversion of sample DNA
- Backbone of EpiDirect® methylation qPCR assays
- Enables direct qPCR-based quantification of DNA methylation
Product Details
Reference Numbers:
670-671/672-673
Product Name:
EpiDirect® Anchor; PentaGreen-Green Quencher/EpiPrimer™
Purification:
HPLC
Yield:
10/50 nmol or higher delivered
Format:
Lyophilized or dissolved in water/TE buffer
Verification:
Melt analysis of EpiDirect® Anchor using complementary methylated and unmethylated target
Publications
A qPCR technology for direct quantification of methylation in untreated DNA
Bendixen et al. – Nature Commun. – 2023
Related Files
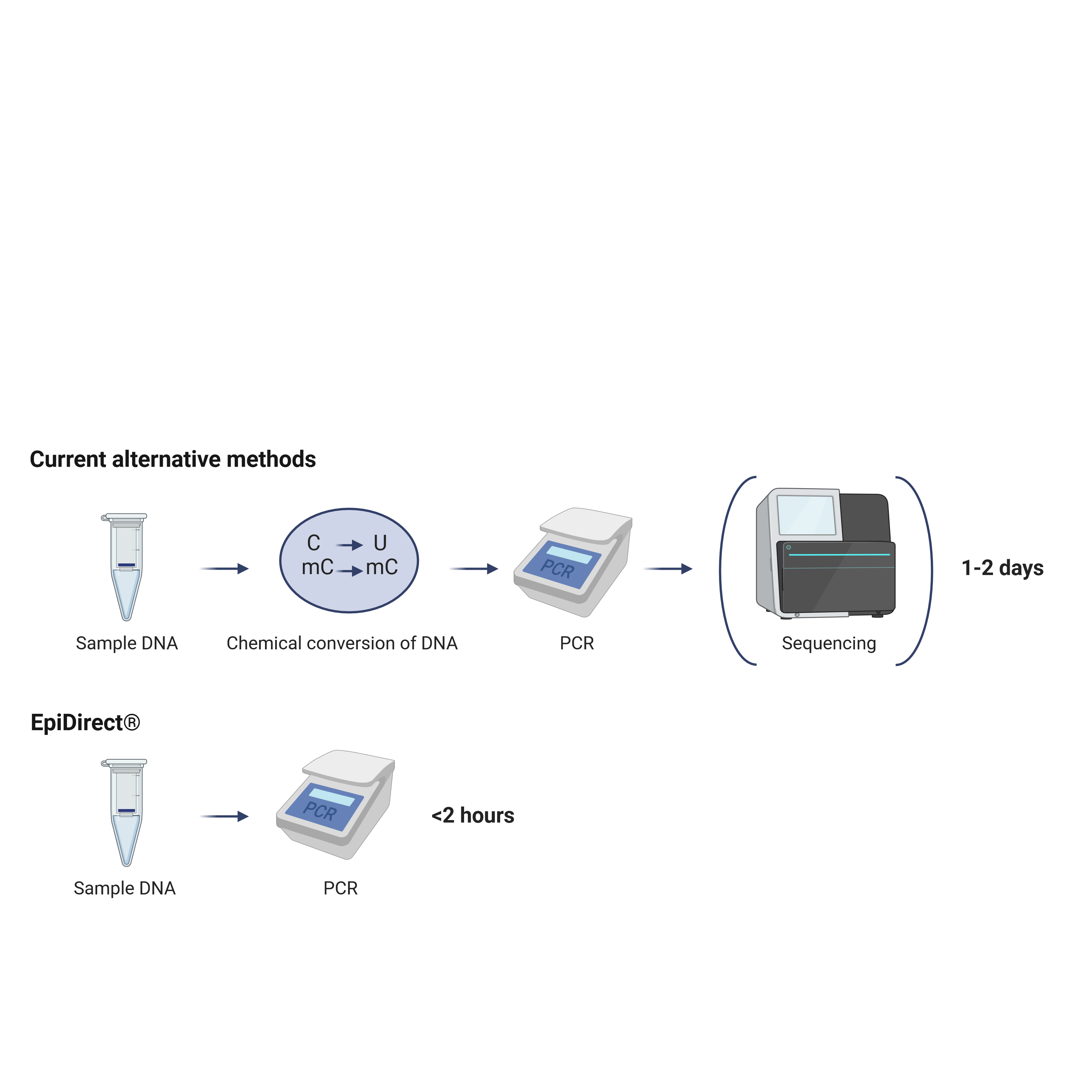
EpiPrimer™ Design and Principle
The EpiPrimer™ is the technological backbone of the EpiDirect® assays and is designed to bind with high affinity to the methylated target and thereby specifically prime direct amplification of methylated DNA vs. non-methylated DNA without prior chemical conversion.
The EpiPrimer™ consists of 3 parts: a) an IPN-containing anchor sequence that binds to the methylated target region, b) a starter sequence that primes DNA replication, and c) a loop sequence that is part of the subsequent methyl-unspecific priming. Thus, the anchor and starter sequence prime the initial replication of the methylated DNA while the loop and starter sequence combined work as a primer in the remaining PCR cycles.
When combined with a methylation-independent reference assay, the methylation-dependent EpiPrimer™-based assay can be used to determine the methylation frequency af the EpiPrimer™ target region.
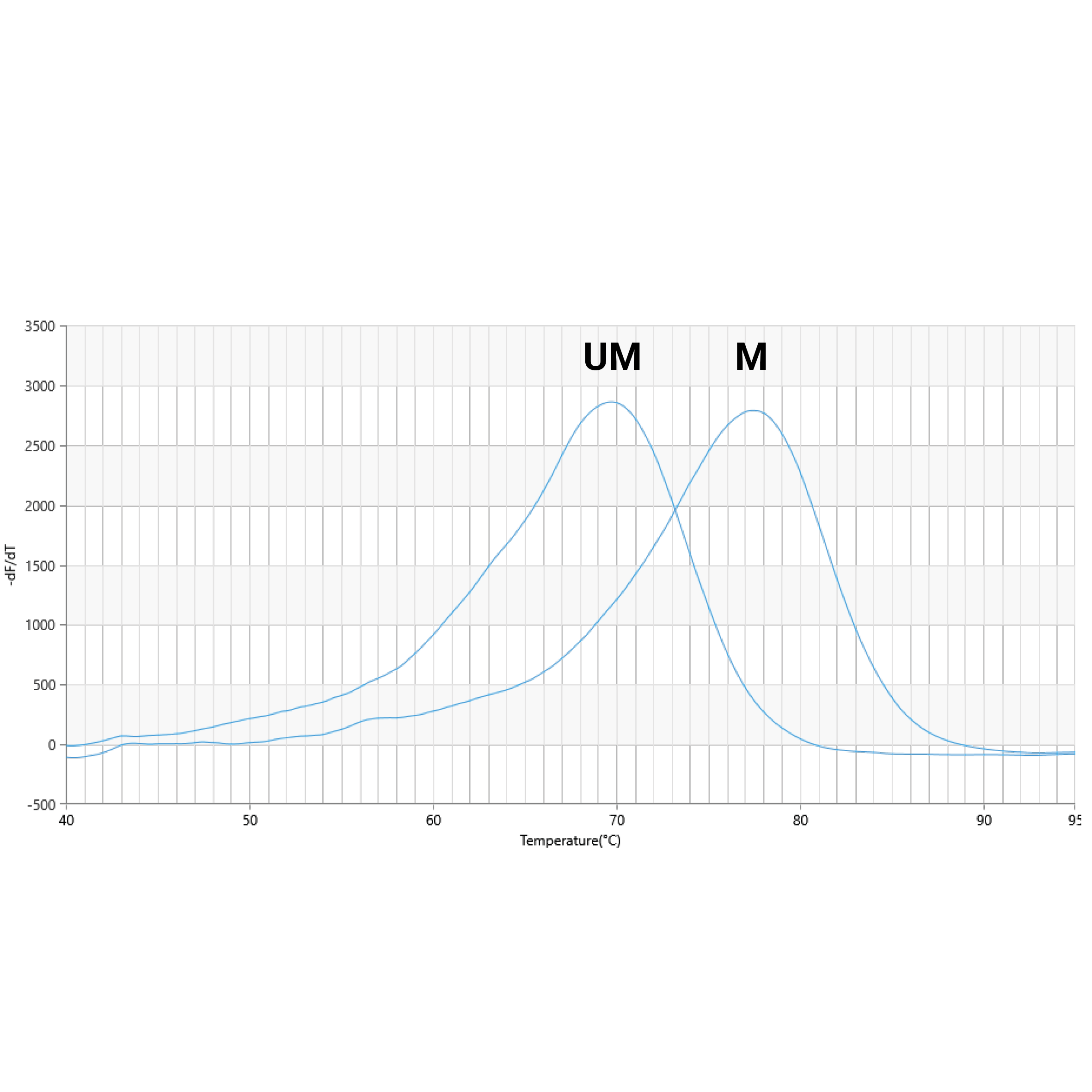
Functional Verification
As part of our quality control, the anchor sequence of the EpiPrimer™ is verified by melt analysis using both methylated (M) and unmethylated (UM) target. A minimum melting temperature difference of 5°C is guaranteed for anchors designed by PentaBase.

EpiDirect® Design Services
PentaBase offers different services related to design and verification of custom Epidirect® oligos and complete assays. Reach out to us if you are interested in developing customised EpiDirect® assays.